This will be the last Microbial Advent Calendar of the season. Today we will in the most biased way, highlight one of the best host-microbe association models we can currently work with. I am talking about the nematode Caenorhabditis elegans. This is a fascinating system, both by its history and its ease of use.
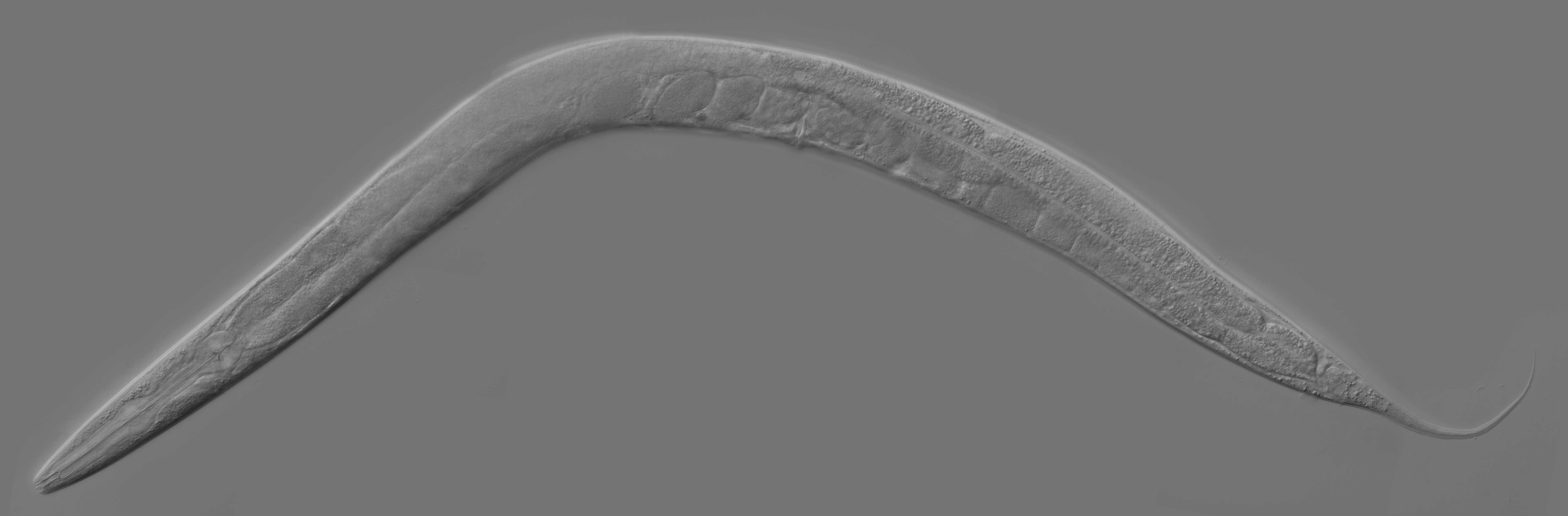
First described by the French zoologist Emile Maupas in 1897 when he discovered two worms in humus soil around Alger. From there this nematode was thought to be living in soils. Way later in 1963 Sydney Brenner proposed to use C. elegans as a model organism for the investigation primarily of neural development in animals. It is one of the simplest organisms with a nervous system and it is easy to maintain in laboratory and culture in very large batches.
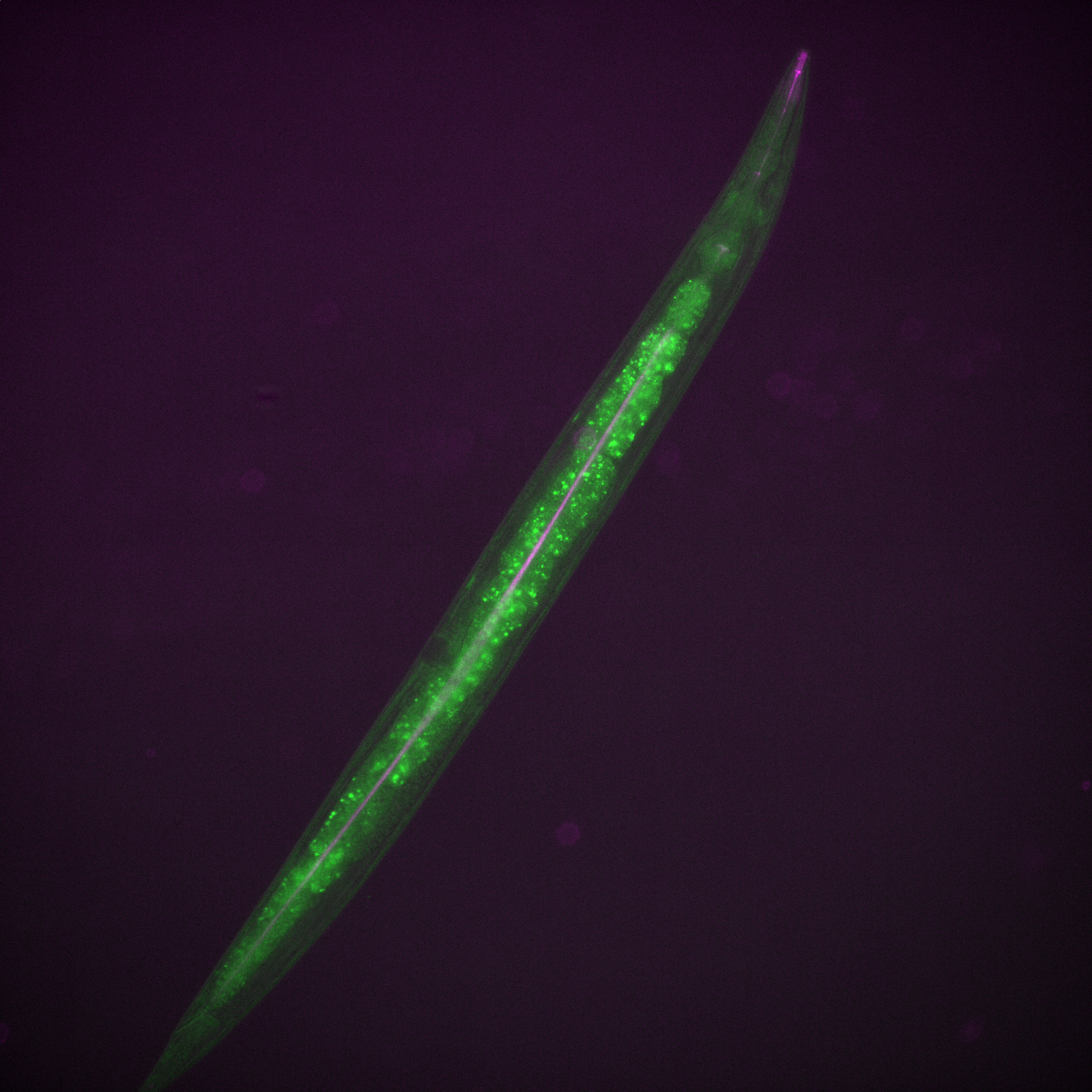
Have a fancy fluorescent worm for no other reason that it’s own awesomeness!
For the past 60+ years, the nematode has been studied in depth and now has a tremendous amount of knowledge database to explore a new aspect of science. One of the truly exciting aspects of this model is that the developmental fate of every single somatic cell (959 in the adult hermaphrodite; 1031 in the adult male) has been mapped. Meaning that we know from the embryo cell which one is going to generate which one in the adult organism!
If C. elegans has been widely used for development, neuronal and behavioral studies there is still a huge knowledge gap about the worm: It’s ecology. Like I mentioned in the first paragraph, it was thought for the longest time to be a soil nematode. However, despite the efforts of many researchers to sample the worm in the wild, it was never found in abundance in soils. What became clear instead was that C. elegans is a colonizer of various microbe-rich habitats, in particular, decaying plant matter. You will then find it on compost heaps, rotting fruits and other plant stems. When food is scarce, C. elegans disperse via carriers, such as slugs, snails, isopods or myriapods until they find the next nutrient stash.
Another area where we know little is the Caenorhabditis overall phylogeny. Although there are 25 described species of Caenorhabditis. Their natural habitat and ecological specificities are very poorly understood, mainly because many species have been isolated only once or twice. One curious example is C. drosophilae, which seems to be ecologically specialized: this species has been found on rotten Saguaro cactus in Arizona, in a transport association with the fly Drosophila nigrospiracula, allowing dispersal between cactus plants. If all of those Caenorhabditis are morphologically similar, they are highly divergent at the genetic level. For the longest time, people have been looking for a C. elegans sister species, to have a close relative to compare to. The search might finally be over this year with the description of C. inopinata found on fig tree leaves in Japan (paper here).
You can find good papers reviewing the state of knowledge of C. elegans ecology here and here.
You can guess that if the ecology of the worm was a little bit overlooked in the past decades, something similar happened to its feeding habits and biology. Like does the worm have a microbiome? Well, turns out it does! After 60 years of development studies, multiple labs went back into the wild to look at what type of worms are out there and what bacteria they are associated with. It resulted in a series of papers showing that indeed they are associated with specific bacterial communities (reviewed here and here).
C. elegans used for microbiome studies revealed a new powerful aspect of the model system. By default, and for a long time, C. elegans work was done in the one Bristol N2 strains. A strain of C. elegans carried over and over and over in the laboratory on the same strain of bacteria (Escherichia coli OP50). But this is not the only strain of the worm, hundreds of strains isolated from the wild are available and have a phenotype which as not yet evolved to be laboratory tamed. This provides on one side a huge pool of different hosts to study what is the effect of natural genetic variation on the microbiome association. On the other side, because you can produce gnobiotic worms, you can finely control the microbiome composition by feeding known bacteria. Both sides have genomic editing tools available which make the whole system the perfect sandbox/playground for the molecular biologists looking to dissect host-microbe interactions.
My reaction realizing how cool it is to work with C. elegans